Solving the 3D structure of a protein from amino acid sequences has been the mystery that has puzzled many scientists. With the advancement in AI, a biologist at Harvard have developed an algorithm which he claimed it will work 1 million times faster than Google’s
AlphaFold, fastest protein structure predictor for the moment.
Prediction of Protein structure using the amino acid sequence has been the grandest quest, biologist of the world is trying to solve. The race to predict the structure from the amino acid sequence has intensified in the last decade. Many scientists using mathematical modeling, molecular biology, and computer science have developed many tools and algorithms to predict the correct structure with great efficiency and less time.
AlphaFold is one of those, an algorithm developed by Google’s firm Deep mind that predicts proteins structure faster than the present techniques by at least 15%.
AlphaFold: The algorithm uses neural networks to predict physical properties. It relied on deep neural networks that
are trained to predict the properties of the protein from its genetic sequence. The properties the networks predict are: (a) the distances between pairs of amino acids and (b) the angles between chemical bonds that connect those amino acids.
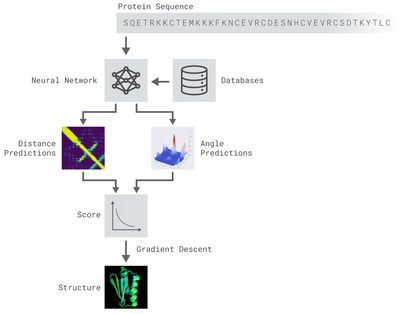 |
| Principle of AlphaFold protein structure predictor |
Critical Assessment of protein Structure Prediction (CASP) is a community-wide, worldwide experiment for protein structure prediction taking place every two years since 1994.
Over 100 research groups from all over the world take part in CASP. In 2018,
Alphafold won the CASP13 outperforming all the contenders by almost 15%.
This year a biologist from Harvard, Mohammed
AIQuraishi has created an algorithm he claimed will work 1 million times faster than the
AplhaFold. “
AIQuraishi’s approach is very promising,” said lan Holmes, a computational biologist at the University of California, Berkley.
His algorithm
is inspired by the brain’s wiring that learns from examples. It’s fed with known data on how amino acids sequences map to protein structure and then learn to produce a new structure from unfamiliar sequences.
References:
1.
AlQuraishi, M. Cell Syst. 8, 292–301, 2019.
Facebook Layman's Biology
Twitter
LaymanBiology

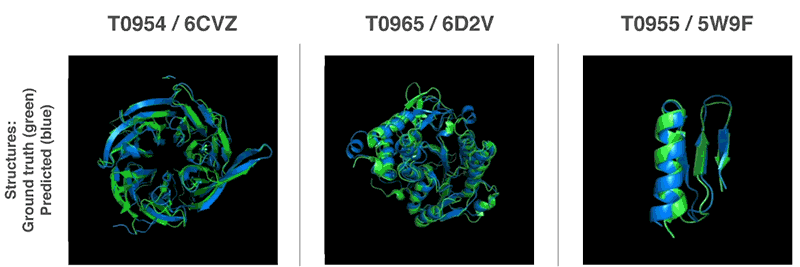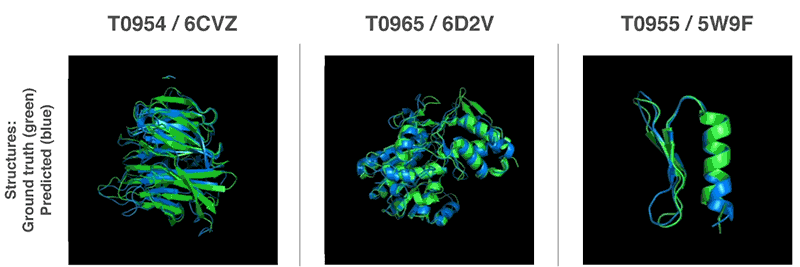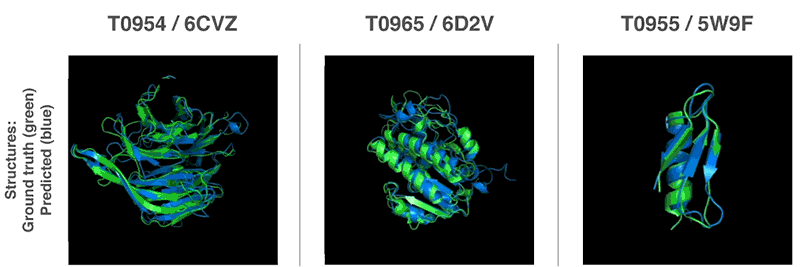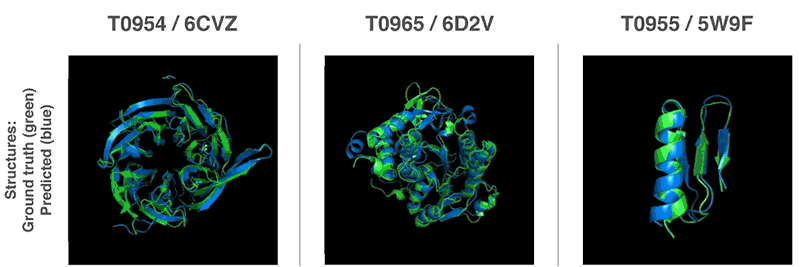
Comments
Post a Comment
Leave your comment about the post